|
He Group Projects
This web page introduces the research projects conducted in the Hegroup:
(1) Development and applications of community-based interoperable ontologies
Ontologies have emerged to become critical to biomedical data and knowledge integration, sharing and validation, as well as new knowledge discovery. A major field of AI is knowledge representation and reasoning (KR², KR&R), for which ontology is a part of the story and critical.
We have initiated and led the development of many community-based ontologies such as the Cell Line Ontology (CLO), Ontology of Biological and Clinical Statistics (OBCS), and Ontology of Host-Microbiome Interactions (OHMI). Dr. He is also an active developer of the Ontology for Biomedical Investigations (OBI), which is collaboratively developed by over 20 communities, and Dr. He is the representation of the vaccine community. To support the Kidney Precision Medicine Project, Dr. He is leading the development of the Kidney Tissue Atlas Ontology (KTAO). To face the challenge of the pandemic COVID-19, Dr. He has initiated the development of the community-driven Coronavirus Infectious Disease Ontology (CIDO).
These ontologies can be used in many applications, including standardized data and knowledge representation, sharing, integration, and advanced analysis. We can also apply ontology to different informatics methods such as literature mining and machine learning.
Representative publications:
- Sarntivijai S, Lin Y, Xiang Z, Meehan TF, Diehl AD, Vempati UD, Schürer TC, Pang C, Malone J, Parkinson H, Liu Y, Takatsuki T, Saijo K, Masuya H, Nakamura Y, Brush MH, Haendel MA, Zheng J, Stoeckert CJ, Peters B, Mungall CJ, Carey TE, States DJ, Athey BD, He Y. CLO: The Cell Line Ontology. Journal of Biomedical Semantics. 2014, 5:37. doi:10.1186/2041-1480-5-37. PMID: 25852852. PMCID: PMC4387853.
- Yongqun He Y, Sirarat Sarntivijai, Yu Lin, Zuoshuang Xiang, Abra Guo, Shelley Zhang, Desikan Jagannathan, Luca Toldo, Cui Tao and Barry Smith. OAE: The Ontology of Adverse Events. Journal of Biomedical Semantics. 2014, 5:29 doi:10.1186/2041-1480-5-29. PMID: 25093068.PMCID: PMC4120740.
- Zheng J, Harris MR, Masci AM, Lin Y, Hero A, Smith B, He Y. The Ontology of Biological and Clinical Statistics (OBCS) for Standardized and Reproducible Statistical Analysis. J Biomed Semantics. 2016. Sep 14;7(1):53. PMID: 27627881. PMCID: PMC5024438.
- Xie J, Zhao L, Zhou S, He Y. Statistical and ontological analysis of adverse events associated with monovalent and combination vaccines against hepatitis A and B diseases. Scientific Reports. 2016. 6:3418. PMID: 27694888. PMCID: PMC5046117.
- He Y, Wang H, Zheng J, Beiting DP, Masci AM, Yu H, Liu K, Wu J, Curtis JL, Smith B, Alekseyenko AV, Obeid JS. OHMI: the ontology of host-microbiome interactions. J Biomed Semantics. 2019 Dec 30;10(1):25. PMID: 31888755. PMCID: PMC6937947.
- Note: check all Dr. He's ontology-related research papers.
The following is a list of ontologies that we have been leading in development. Note that these ontology development all follows the OBO Foundry principles and uses BFO as the upper level ontology:
 |
Ontology of Adverse Events (OAE): A community-based ontology targeting to represent adverse events for drugs, vaccines, and other biomedical reagents. Note: OAE was previously named AEO. (Reference: PMID: 25093068). |
 |
Cell Line Ontology (CLO): A community-based cell line ontology. The CLO development originated from the research done by Sirarat (Sira) Sarntivijai before she joined He Lab for PhD thesis research. Sira got her PhD with ontology-focused research in 2012. Sira is still actively working on CLO update with international collaborations. (Reference: http://www.jbiomedsem.com/content/5/1/37/). |
 |
Ontology of Genes and Genomes (OGG): A community-driven ontology of genes and genomes. (Reference: ICBO 2014 full length proceeding paper). |
 |
Interaction Network Ontology (INO): an ontology in the domain of interaction networks. INO develoment follows the OBO Foundry principles. INO can be used for automated reasoning such as literature mining and Bayesian network reasoning. (References: JBMS INO paper; arXiv:1311.3355). |
 |
Vaccine Ontology (VO): The VO project is a community-based project with an aim to develop the VO to ensure vaccine data standardization, exchange, and automated reasoning. (Reference: Nature Procedings 2009; PMID: 23256535; PMID: 21624163; PMID: 24209834) . |
Other ontologies not listed above:
Note: Oliver prepared two tutorial websites. Although not routinely updated, they may still be helpful:
(2) Development of Ontoanimal tools to support interoperable ontology development & applications
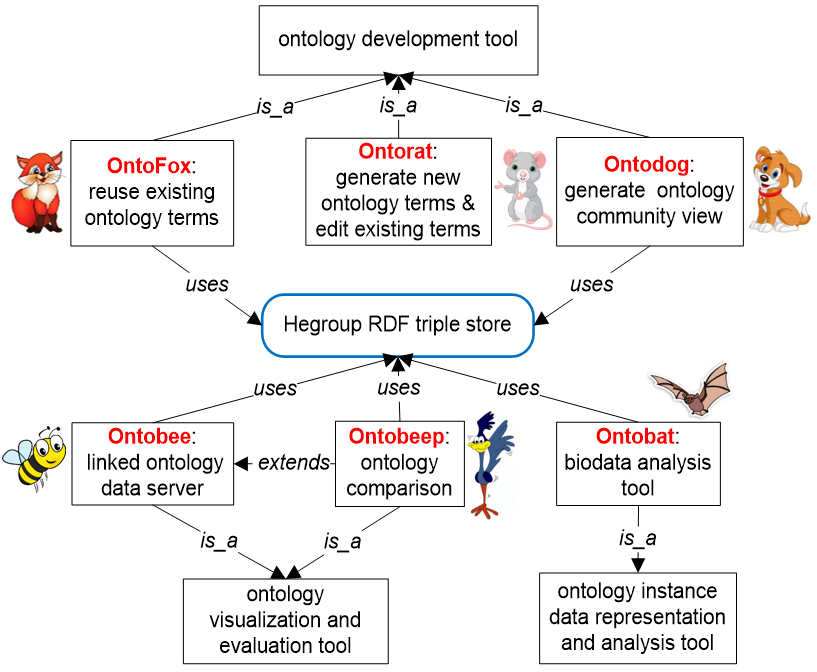
We have developed many widely used ontology tools (e.g., Ontobee, Ontofox, and Ontorat), collectively names “Ontoanimal” tools. Each tool has specific functions; together, these tools are able to extract ontology subsets, provide ontology community views, generate and edit ontology terms, query and visualize ontology terms, provide statistics of ontologies, and compare ontologies.
Representative publications:
- Xiang Z, Courtot M, Brinkman RR, Ruttenberg A, He Y. OntoFox: web-based support for ontology reuse. BMC Research Notes. 2010, 3:175. PMID: 20569493. PMCID: PMC2911465.
- Zheng J, Xiang Z, Stoeckert Jr. CJ, He Y. Ontodog: a web-based ontology community view generation tool. Bioinformatics. 2014, 30(9):1340-2. doi: 10.1093/bioinformatics/btu008. [PDF] PMID: 24413522. PMCID: PMC3998133.
- Xiang Z, Zheng J, Lin Y, He Y. Ontorat: Automatic generation of new ontology terms, annotations, and axioms based on ontology design patterns. Journal of Biomedical Semantics. 2015, 6:4 doi:10.1186/2041-1480-6-4. PMID: 25785185. PMCID: PMC4362828.
- Ong E, Xiang Z, Zhao B, Liu Y, Lin Y, Zheng J, Mungall C, Courtot M, Ruttenberg A, He Y. Ontobee: A linked ontology data server to support ontology term
dereferencing, linkage, query, and integration. Nucleic Acid Research. 2017 Jan 4;45(D1):D347-D352. PMID: 27733503. PMCID: PMC5210626.
- He Y, Xiang Z, Zheng J, Lin Y, Overton JA, Ong E. The eXtensible ontology development (XOD) principles and tool implementation to support ontology interoperability. J Biomed Semantics. 2018 Jan 12;9(1):3. PMID: 29329592. PMCID: PMC5765662.
Below is a quick summary of these Ontoanimal tools.
 |
Ontobee: A Linked Data Server that publishes RDF and HTML data simultaneously. The Ontobee web server aimed to facilitate ontology visualization, query, and development. OntoBee provides a user-friendly web interface for displaying the details and its hierarchy of a specific ontology term. Meanwhile, OntoBee provides a RDF source code for the particular web page, which supports remote query of the ontology term and the Semantic Web. (Reference: Xiang et al., 2011). |
 |
OntoFox: An Ontology tool that Fetches ontology terms and axioms. OntoFox is a web-based system to support ontology reuse. OntoFox facilitates ontology development by automatically fetching ontology terms and their annotations from existing ontologies and saving the results in an importable RDF/OWL format. (Reference: PMID: 20569493). |
 |
Ontodog: A web-based ontology community view generator. (Reference: PMID: 24413522). |
 |
Ontorat: An ontology tool that supports Ontology representation of axioms using templates and design patterns. Ontorat is a web server that automatically generates new ontology terms and axioms based on user-provided input file. (Reference: Xiang et al, 2012). |
It is noted that most of our ontology tools use our Hegroup Resource Description Framework (RDF) triple store. The SPARQL endpoint of our triple store is: http://sparql.hegroup.org/sparql/.
We have also initiated the proposal of the eXtensible ontology development (XOD) principles and methods to support ontology interoperability. The above mentioned Ontoanimal tools can be used to support the implementation of the XOD principles:
With the XOD and our Ontoanimal tools, ontology development is no longer boring and has become more fun and useful!
(3) Vaccine informatics, host-pathogen interactions, and vaccine/drug safety
We study Vaccine Informatics, a branch of vaccinology that Dr. He and his colleagues pioneered. We have developed VIOLIN, the most comprehensive vaccine database and analysis system. The Vaccine Ontology (VO) has become a community effort for standard representation of vaccines, vaccine components, vaccination, and host responses to vaccines. We have also developed Vaxign, the first web-based publically available vaccine target design tool based on bioinformatics analysis of genome sequences using the strategy of reverse vaccinology.
We study host-pathogen interactions. We developed PHIDIAS, the most comprehesenive host-pathogen interaction database focusing on human and animal pathogens.
As part of PHIDIAS, we have developed Victors, a comprehensive knowledge base of more than 3,000 virulence factors in >120 human and animal pathogens. We have also developed an Ontology of Host-Pathogen Interactions (OHPI) to logically represent the virulence factors and their interactions with the hosts.
We are interested in vaccine and drug safety study.
We have led the development of the community-based Ontology of Adverse Events (OAE), and applied it to study vaccine and drug adverse events.
Vaccine Informatics. We have developed the VIOLIN vaccine database and analysis system. As a part of VIOLIN, we have developed Vaxign, the first web-based publically available vaccine target design tool based on bioinformatics analysis of genome sequences using the strategy of reverse vaccinology. logy. Comparative genomics. Various genome sequences can be compared to address many biomedical questions. Analysis of vaccine and drug adverse event mechanisms. We are interested in applying the Ontology of Adverse Events to study the mechanisms of vaccine and drug-induced adverse events.
Representative publications:
- Xiang Z, Tian Y, He Y. PHIDIAS: a pathogen-host interaction data integration and analysis system. Genome Biology. 2007 Jul 30;8(7):R150. PMID: 17663773. PMCID: PMC2323235.
- He Y, Xiang Z, Mobley HLT. Vaxign: the first web-based vaccine design program for reverse vaccinology and an application for vaccine development. Journal of Biomedicine and Biotechnology. Volume 2010 (2010), Article ID 297505, 15 pages. [PMID: 20671958]. PMCID: PMC2910479.
- Racz R, Chung M, Xiang Z, and He Y. Systematic annotation and analysis of “virmugens” - virulence factors whose mutants can be used as live attenuated vaccines. Vaccine. 2013 Jan 21;31(5):797-805. Epub Dec. DOI: 10.1016/j.vaccine.2012.11.066. PMID: 23219434. PMCID: PMC3556277.
- Sayers S, Li L, Ong E, Deng S, Fu G, Lin Y, Yang B, Zhang S, Fa Z, Zhao B, Xiang Z, Li Y, Zhao Z, Olszewski MA, Chen L, He Y. Victors: a web-based knowledge base of virulence factors in human and animal pathogens. Nucleic Acid Research. 2019 Jan 8;47(D1):D693-D700. doi: 10.1093/nar/gky999. PMID: 30365026.
PMCID: PMC6324020.
- Ong E, Wang H, Wong MU, Seetharaman M, Valdez N, He Y. Vaxign-ML: supervised machine learning reverse vaccinology model for improved prediction of bacterial protective antigens. Bioinformatics. In press (Acceptance date: 2020-02-18). DOI: 10.1093/bioinformatics/btaa119. PMID: 32096826.
Related tools developed in He group:
 |
VIOLIN: Vaccine Investigation and Online Information Network: A web-based database system for research and development of vaccines against various human pathogens (e.g., HIV, influenza, and tuberculosis) with high priority in public health and national biological defense. |
 |
PHIDIAS: Pathogen-Host Interaction Data Integration and Analysis System: A web-based data integration and analysis system for biomedical researchers to investigate genomic sequences, curated literature information, and gene expression data related to pathogen-host interactions for those pathogens with high priority in public health. |
Other tools not listed above:
- Vaxign, the first web-based reverse vaccinology tool for vaccine design
- Victors, a comprehensive knowledge base of virulence factors in human and animal pathogens.
- BBP (Brucella Bioinformatics Portal): A gateway for Brucella researchers to search, analyze, and curate Brucella genome data originated from public databases and literature.
(4) Systematic ontology and bioinformatics analysis of biological interaction networks
We hypothesized that ontology supports literature mining and analysis of biological interaction networks. As a result, we developed the Interaction Network Ontology (INO) and applied it to enhance literature mining performance. We are also collaborating with our collaborators, including Drs. Junguk Hur and Arzugan Özgür, to apply ontology for more advanced literature mining.
Bayesian network (BN) can model linear, nonlinear, combinatorial, and stochastic relationships among variables across multiple levels of biological organizations. We have developed new BN algorithms and tools for analysis of gene interaction networks using high throughput gene expression data.
Representative publications:
- Xiang Z, Minter RM, Bi X, Woolf PJ, He Y. miniTUBA: medical inference by network integration of temporal data using Bayesian analysis. Bioinformatics. 2007 Sep 15;23(18):2423-32. PMID: 17644819.
- Hodges AP, Dai D, Xiang Z, Woolf P, Xi C, He Y. Bayesian network expansion identifies new ROS and biofilm regulators. PLoS ONE. 2010 Mar 3;5(3):e9513. PMID: 20209085. PMCID: PMC2831072.
- Hur J, Xiang Z, Feldman EL, He Y. Ontology-based Brucella vaccine literature indexing and systematic analysis of gene-vaccine association network. BMC Immunology. 12(1):49 2011 Aug 26. PMID: 21871085. PMCID: PMC3180695.
- Xiang Z, Qin T, Qin Z, He Y. A genome-wide MeSH-based literature mining system predicts implicit gene-to-gene relationships and networks. BMC Systems Biology. 2013, 7(Suppl 3):S9. doi:10.1186/1752-0509-7-S3-S9. (note: this paper was also presented in InCoB2013 and collected in F1000Posters; see the presentation slides) PMID: 24555475. PMCID: PMC3852244.
- Hur J, Özgür A, Xiang Z, He Y. Development and application of an Interaction Network Ontology for literature mining of vaccine-associated gene-gene interactions. Journal of Biomedical Semantics. 2015, 6:2. doi:10.1186/2041-1480-6-2. PMID: 25785184. PMCID: PMC4362819.
- He Y. Ontology-based vaccine and drug adverse event representation and theory-guided systematic causal network analysis toward integrative pharmacovigilance research. Current Pharmacology Reports. 2016, Volume 2, Issue 3, pp 113-128. DOI: 10.1007/s40495-016-0055-0. PMID: 27458549. PMCID: PMC4957564.
Related tools developed in He group:
 |
miniTUBA: Medical Inference by Network Integration of Temporal Data Using Bayesian Analysis: A web-based dynamic Bayesian analysis engine for clinical and biomedical researchers to perform medical inference and predictions with temporal datasets. |
 |
GenoMesh: A genome-wide analysis of gene-to-gene relationships and pathways based on the association between individual genes and MeSH terms obtained from literature. Currently GenoMesh includes data on Escherichia coli and Brucella spp.. |
Other tools not listed above:
- Ignet (Integrated gene network), a centrality- and ontology-based liteature discovery system for analyzing and visualizing biological gene interaction networks using all PubMed literature papers.
- Dignet, a program inside Ignet that is able to dynamically search PubMed papers.
- CRCView: A web-based microarray data analysis and visualization system, powered by a model-based clustering algorithm Chinese Restaurant Cluster (CRC) developed by Dr. Steve Qin. CRCView also incorporates several Bioconductor microarray analysis programs including GOStats, genefilter, and Heatplus.
In a 2014 paper (He, 2014) and then a 2016 paper (He, 2016), Dr. He proposed a new “OneNet Theory of Life”. The OneNet theory states that the whole process of a life of an organism is a single complex and dynamic network (called “OneNet”).
Dr. He also proposes OneNet tenets to characterize different aspects of the OneNet life. Ontologies and ontology-based bioinformatics tools, such as those introduced above, can be used to integratively represent and study such OneNet theory and OneNet knowledge of different organisms.
Studying the theory has become Dr. He's hobby.
(5) Wet-lab microbiology and immunology research (not active now):
Our primary biomedical research interest is to understand pathogenesis and immunology of infectious diseases and develop vaccines against intracellular pathogens. In particular, we study Brucella, a facultative intracellular bacterium that causes zoonotic brucellosis in humans as well as domestic and wild life animals. Our research focuses on the role of Caspase-2, an enzyme that is critical in regulating cell death, DNA damage, stress, cancer, and microbial infections. Our research aims to continuously elucidate the caspase-2-mediated proinflammatory cell death pathway and its biological effect on microbial pathogenesis and protective immunity against brucellosis and other diseases.
Our wet-lab research is focused on the study of Brucella, a facultative intracellular bacterium that causes brucellosis, one of the most common zoonotic diseases in the world in humans and a variety of animal species. Our wet-lab Brucella research focuses on the analysis of caspase-2-mediated cell death and its role in Brucella pathogenesis and immunity. Our studies first identified a new caspase-2-mediated proinflammatory cell death, which exists in macrophages and dendritic cells infected with live attenuated rough Brucella strains (e.g., B. abortus cattle vaccine RB51 and B. suis vaccine candidate VTRS1) (see references: Chen & He, 2009, Chen et al, 2011, Li & He, 2012, and Bronner, O'Riordan, and He, 2013). This type of cell death is different from non-proinflammatory apoptosis or caspase-1-mediated proinflammatory pyroptosis, and so we named the cell death "caspase-2-mediated pyroptosis". Interestingly, virulent Brucella inhibits such cell death in infected macrophages (He et al, 2006) but not in dendritic cells (Li & He, 2012). Based on the study of the RB51-macrophage interaction model, endoplasmic reticulum stress activates the inflammasome via NLRP3- and caspase-2-driven mitochondrial damage (Bronner et al, 2015).
Representative publications:
- He Y, Reichow S, Ramamoorthy S, Ding X, Lathigra R, Craig JC, Sobral BW, Schurig GG, Sriranganathan N, Boyle SM. Brucella melitensis triggers time-dependent modulation of apoptosis and down-regulation of mitochondria-associated gene expression in mouse macrophages. Infection and Immunity. 2006 Sep;74(9):5035-46. PMID: 16926395. PMCID: PMC1594834.
- Chen F, He Y. Caspase-2 mediated apoptotic and necrotic murine macrophage cell death induced by rough Brucella abortus. PLoS ONE. 2009 Aug 28;4(8):e6830. PMID: 19714247. PMCID: PMC2729395.
- Chen F, Ding X, Ding Y, Xiang Z, Li X, Ghosh D, Schurig GG, Sriranganathan N, Boyle SM, He Y. Proinflammatory caspase-2 mediated macrophage cell death induced by rough attenuated Brucella suis. Infection and Immunity. 2011 Jun;79(6):2460-9. PMID: 21464087. PMCID: PMC3125819.
- Li X, He Y. Caspase-2-dependent dendritic cell death, maturation, and priming of T cells in response to Brucella abortus infection. PLoS ONE, 2012, 7(8): e43512. doi:10.1371/journal.pone.0043512. PMID: 22927979. PMCID: PMC3425542.
- Bronner DN, O'Riordan MXD, He Y. Caspase-2 mediates a Brucella abortus RB51-induced hybrid cell death having features of apoptosis and pyroptosis. Frontiers in Cellular and Infection Microbiology. 2013. 3:83. PMID: 24350060. PMCID: PMC3842122.
- Bronner DN, Abuaita BH, Chen X, Fitzgerald KA, Nunez G, He Y, Yin XM, O’Riordan MXD. ER stress activates the inflammasome via NLRP3-caspase-2 driven mitochondrial damage. Immunity. 2015 Sep 15;43(3):451-62. PMID: 26341399. PMCID: PMC4582788.
- Note: check all Dr. He's Brucella research papers.
We further hypothesize that caspase-2-mediated immunity is important not only to Brucella, but also to other pathogens. We had experimentally tested how caspase-2 mediates the immune responses of macrophages infected by tuberculosis vaccine BCG, and generated some good prototype results. However, we have not continued this line of research. Instead, we have focused our research on computational medicine and bioinformatics.
Your suggestions, comments, and collaborations are welcome and appreciated.
|
